Published on March 28, 2025 11:24 AM GMT
TLDR
We do not yet understand all the genetic imprints that play an important role in early embryonic development. Known imprinting disorders (Prader-Willi, Angelman, etc.) however, do seem to cover all the most detrimental imprints, since for all cases of uniparental disomy (having 1 pair of chromosomes from only one parent), we know of either the imprinting disorder or of a phenotypically normal case. One exception is maternal UPD 19, which might be rare because proper imprinting on chromosome 19 is crucial for early development in humans.
Imprinting and germline engineering
Most strong methods of germline engineering require us to know what the proper epigenomics of an embryo look like. Measuring the epigenome is quite tricky. Maintaining specifically imprinting is even more tricky. We currently have no method of culturing naive embryonic stem cells that does not lead to loss of imprinting.[1] While there is a lot of literature on imprinting and experiments in mice to determine the mechanisms behind known imprints, research in this area does not happen with the explicit goal of germline engineering. There is no comprehensive list of all human imprints.[2] So there might be regions whose importance for germline engineering in humans might be overlooked if there is no known associated disorder in humans. Hence, this post.
This post assumes basic familiarity with epigenetics. The next paragraph is a 1-paragraph summary, but I recommend reading these two excellent posts on epigenetics. If you know what DNA methylation and imprinting are, you can skip the posts.
The short explanation is that DNA methylations are small chemical marks on the DNA that change which proteins bind well to the DNA. Together with other chemical marks on histones, which are spindles that DNA is rolled around, these marks are how a cell knows that it is a skin cell, rather than a neuron. Imprinting is a phenomenon where the two sets of chromosomes in early mammal embryos behave differently depending on if they were inherited from the father or the mother side. Imprinting appears to be mostly due to differences in DNA methylation and histone marks.
Imprinting disorders and uniparental disomy
One of the earliest ways we discovered imprinting is through disorders like Prader-Willi syndrome, which had already been linked to deletions of some genes on Chromosome 15. Some patients though did not have any such deletions. Instead, they had inherited both of their homologous chromosomes 15 from their mother. This is usually called maternal uniparental disomy 15 (UPD). Uniparental disomies can develop through a few different mechanisms that I'll explain below.
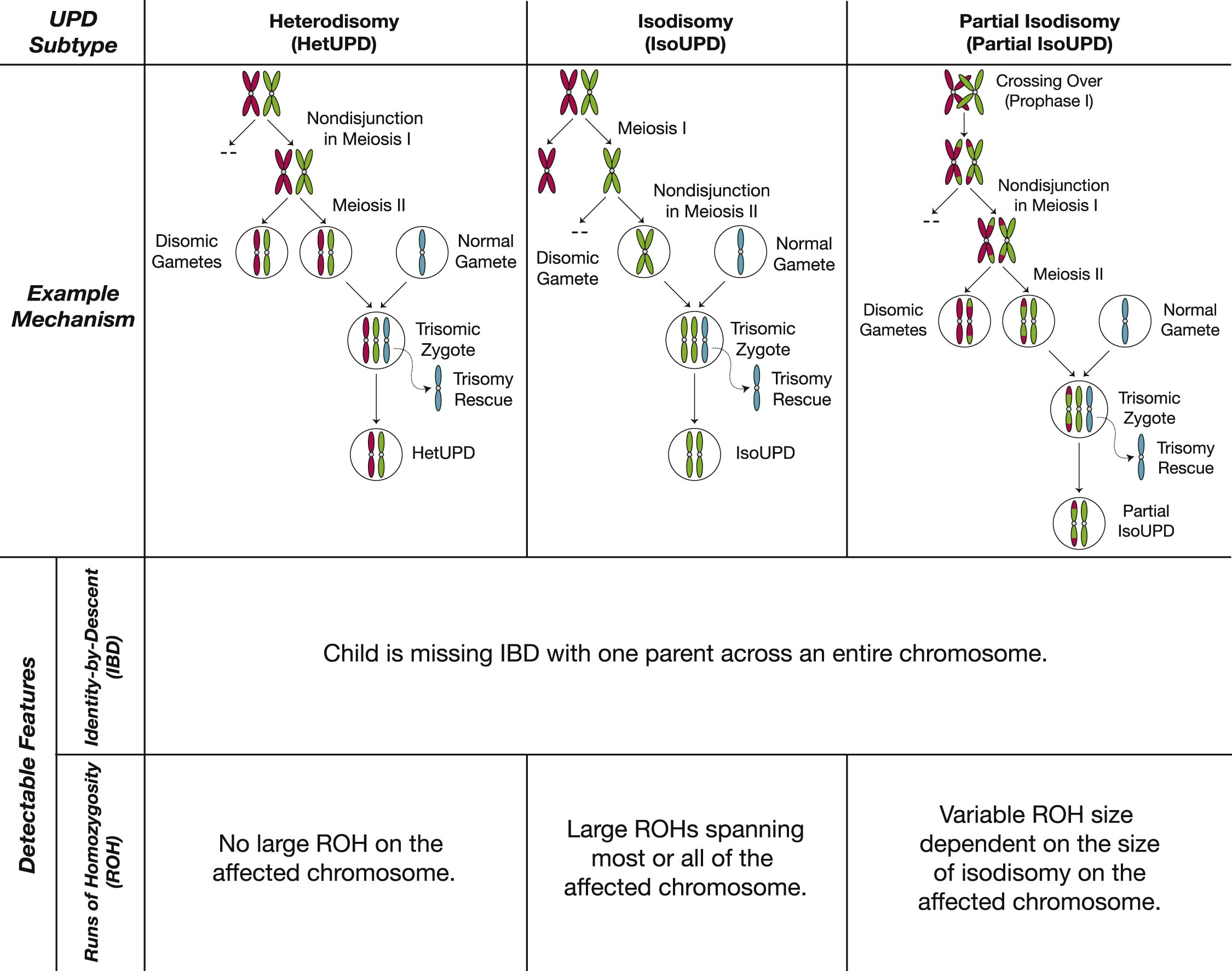
What is uniparental disomy, and how does it develop?
Uniparental disomy means someone has a chromosome twice from the same parent, rather than 1 copy from each parent. In principle, this is an excellent way to identify problems that could be caused by imprinting. Unfortunately, of course, there are additional confounders other than imprinting on the chromosome in UPD. In most cases of UPD, all or parts of the two chromosomes are homozygous. They are respectively called isoUPD and partial isoUPD. This often leads to deleterious recessive genes surfacing, just like with incest.
20% of the time[3] however, UPD comes with heterozygous chromosomes, so the affected chromosomes are just the same as those of one of the parents (or the same chromosomes, but crossed over). Even in those cases, UPD is still not a perfect "treatment group" for the effects of imprinting, since UPDs are most often the result of "rescued" trisomies: trisomies that get accidentally corrected by an error during chromosome segregation, turning one cell disomic and the other one tetrasomic for that specific chromosome. The more healthy disomic cells then outcompete the aneuploid cell, such that the embryo is completely or mostly made up of euploid cells (aneuploid cells tend to form the placenta instead). Analogously, UPD can be the product of a rescued monosomy[4]. In practice, this means, that when looking at a cohort of UPD patients, any symptoms they have in common could also be due to trisomy or monosomy in the placenta or even in some tissues of the patient.[5]
More on known imprinting disorders
Most imprints seem to have evolved due to parental conflict about how much effort to invest in particular offspring in non-monogamous species.[6][7] If the father and mother favour different levels for the expression of a gene and the respective benefits outweigh the benefits of expressing the gene from both chromosomes, then the gene evolves to become imprinted and is only expressed from the side that favours its expression.[7]
This also means every imprinting disorder like Prader-Willi syndrome (caused by maternal UPD) has a sister syndrome that is caused by inheriting both chromosomes 15 from one's father (paternal UPD): Angelmann syndrome.
Through UPD, we later discovered imprinting for a few other chromosomes. Most notably, Silver-Russel syndrome can be caused by maternal UPD on chromosomes 7, 11 or 20. Paternal UPD11 leads to Beckwith-Wiedemann syndrome. Maternal UPD14 leads to Temple syndrome, while Paternal UPD14 leads to Kagami-Ogata syndrome. Paternal UPD 6 leads to transient neonatal diabetes mellitus. Paternal UPD20 leads to Pseudohypoparathyroidism type Ib.
In line with the parental conflict theory of imprinting, when I google the symptoms and first-line treatments for these disorders, maternal UPD is associated with hypoglycemia and patients tend to benefit from growth hormone treatments. Meanwhile, paternal UPD is associated with hyperglycemia and lines of treatment seem more varied. Loss of imprinting is also common in some kinds of cancers,[8] so the corresponding types of UPD probably make it more likely to get cancer.
All of these disorders can also be caused by genetic deletions, which allowed us to identify where the imprints for these disorders are located. Except for maternal UPD7, where so far a specific gene or region hasn't been identified.[9] UPD16 sometimes leads to Silver-Russel symptoms, but it is unclear if this is just due to mosaic trisomy 16 or other confounders.[9]
It turns out, that after these more common cases, uniparental disomy cases are a lot less often reported.
What is up with the underreported UPD cases?
One question was still unclear to me after looking up all the known imprinting disorders: To what extent do we even know all the "lethal" or highly detrimental imprinting regions? By highly detrimental, I mean something that would not go undetected even if the disease is rare. For example, it could be that one of the rare uniparental disomy cases is so deadly already in early pregnancy that we never observed any cases. We would still have caught any such imprinting regions if they also exist in mice, but for any important imprints missing in mice or if they happen to be less of a problem in mice, UPD would be one of the very few ways we could observe them.
It turns out it is probably the latter (imprinting is more benign for the other cases of UPD, although maternal UPD19 might be an exception). There are two main sources of evidence making me believe this.
The first one is this website maintained by Thomas Liehr who fairly comprehensively gathered together most (all?) existing case studies of UPD. I manually went through all the database entries and for all chromosomes except chromosome 5, I was able to find full UPD cases that were "phenotypically normal" for either paternal or maternal UPD. From looking over the cases of chromosome 5 my impression from the variedness of symptoms was that this was more likely due to recessive genetic disorders and mosaic trisomy 5 rather than due to imprinting.
There are some chromosomes, like Chromosomes 19, where we do have 1 case of UPD19 where the child didn't have apparent problems. We do have evidence that there are a few imprints on chromosome 19 (specifically an imprinting cluster in 19q13.4–19q13.43), whose homologs were first discovered through UPD studies in mice.[10] So there are very likely effects from incorrect imprinting on chromosome 19, though they might be small. Also, while for some chromosomes we have phenotypically normal data for adults (among others chr1, chr21, chr22), for others we only have this reported for newborns.
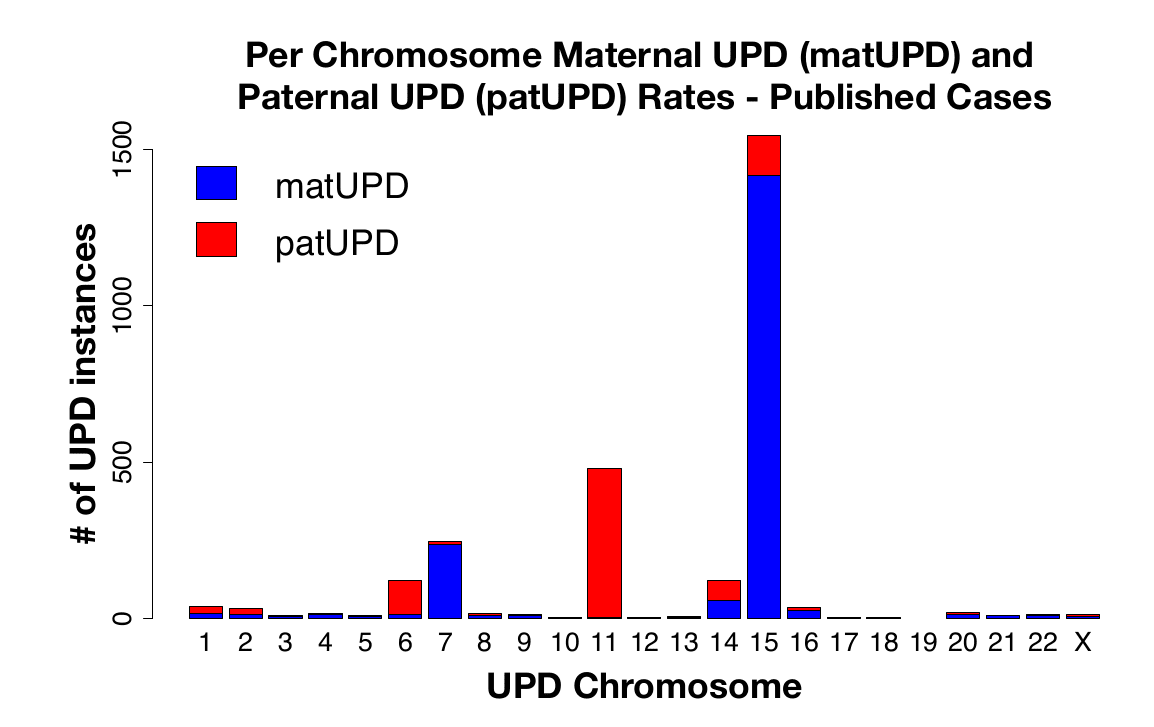
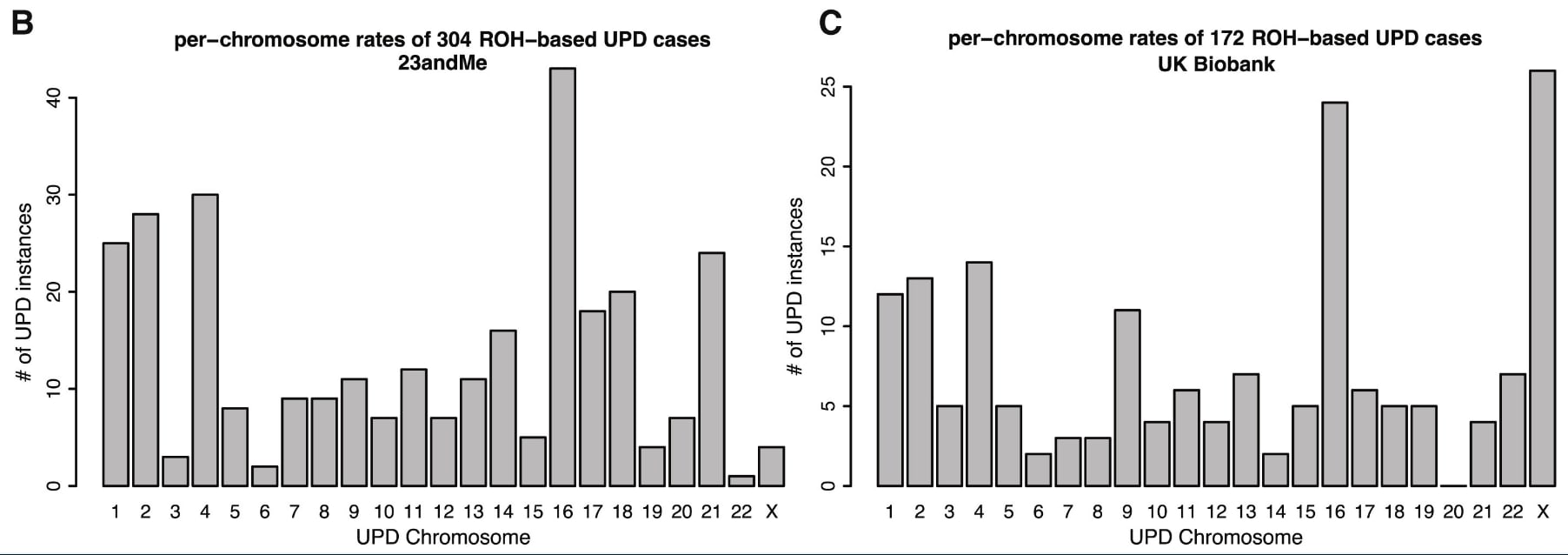
The second line of evidence for less known uniparental disomy cases being benign, comes from this large data analysis of 23andMe's and UK Biobanks data, showing that a lot of these lesser-known imprinting disorders aren't rarer than the commonly reported ones:
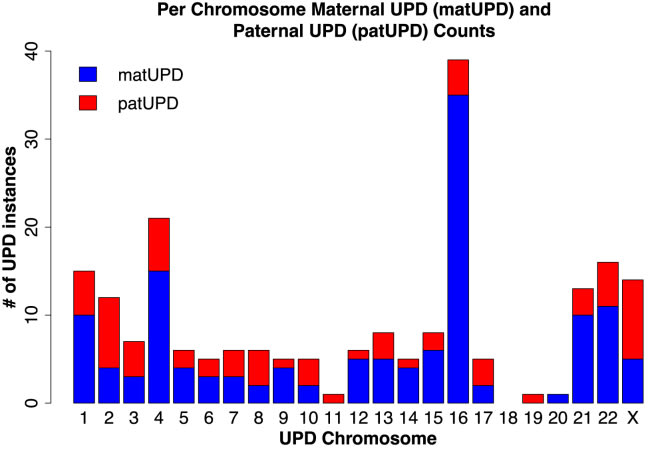
23andMe's dataset has enough parent-child pairs, that we can even get a breakdown by paternal or maternal origin:
If they are this common, we would know the phenotype of UPD5 by now, just as we know it for UPD7, if there was a clear phenotype.
Let's look at the cases, where we don't have maternal and paternal cases in the figure above. In principle, all the above data still makes it possible that maternal UPD19 is quite bad for embryos if all the UPD19 cases in (Nakka et al., 2019) turn out to be paternal UPD19. To my knowledge, there is no reported case of maternal UPD19. So perhaps that imprinting region on chromosome 19 is quite important. Or some other region makes this rare ¯_(ツ)_/¯. UPD18 is not a problem here, since for both maternal and paternal UPD18, there are documented phenotypically normal cases.[11] Both maternal and paternal UPD11 and UPD20 are known to be imprinted, as mentioned earlier.
Conclusion
So overall, while we do not have a complete understanding of all imprints, we have probably identified the most detrimental ones. Exceptions are UPD7, where we do not know which gene(s) causes Silver-Russel syndrome. We are also still in the dark about maternal UPD19, which might be deadly to embryos.
Thanks to Tsvi Benson-Tilsen for discussion and Gene Smith for employing me to do this research.
- ^
Bar, S., Schachter, M., Eldar-Geva, T., & Benvenisty, N. (2017). Large-Scale Analysis of Loss of Imprinting in Human Pluripotent Stem Cells. Cell Reports, 19(5), 957–968. https://doi.org/10.1016/j.celrep.2017.04.020
- ^
The following list is often cited, but a lot of imprints in it are only "predicted": Geneimprint: Genes. (n.d.). Retrieved March 28, 2025, from https://www.geneimprint.com/site/genes-by-species
- ^
See table 1 in [12]
- ^
Rudd, M. K., Schleede, J. B., Williams, S. R., Lee, K., Laffin, J., Pasion, R., & Papenhausen, P. R. (2018). Monosomy X rescue explains discordant NIPT results and leads to uniparental isodisomy. Prenatal Diagnosis, 38(12), 920–923. https://doi.org/10.1002/pd.5349
- ^
Another way UPD can develop is if sperm and eggs are accidentally disomic and nullisomic in the same chromosome, cancelling out each other's mistakes[13][14].
If we get such an accidental UPD case that also happens to be a heterodisomy (presumably 50% of these cases are heterodisomy?) that would be quite close to isolating the effects just due to imprinting and not due to any mosaic trisomie in either the embryo or the placenta. Unfortunately this is not going to happen often.[14]
- ^
There are a lot of other theories how imprinting evolved, but the consensus seems correct on this one. Read this one for more: Spencer, H. G., & Clark, A. G. (2014). Non-conflict theories for the evolution of genomic imprinting. Heredity, 113(2), 112–118. https://doi.org/10.1038/hdy.2013.129
- ^
Moore, T., & Haig, D. (1991). Genomic imprinting in mammalian development: A parental tug-of-war. Trends in Genetics, 7(2), 45–49. https://doi.org/10.1016/0168-9525(91)90230-N
- ^
Jelinic, P., & Shaw, P. (2007). Loss of imprinting and cancer. The Journal of Pathology, 211(3), 261–268. https://doi.org/10.1002/path.2116
- ^
Kurup, U., Lim, D. B. N., Palau, H., Maharaj, A. V., Ishida, M., Davies, J. H., & Storr, H. L. (2024). Approach to the Patient With Suspected Silver-Russell Syndrome. The Journal of Clinical Endocrinology and Metabolism, 109(10), e1889–e1901. https://doi.org/10.1210/clinem/dgae423
- ^
Access via Internet archive: Beechey CV, Cattanach BM, Blake A , Peters J (2008), MRC Mammalian Genetics Unit, Harwell, Oxfordshire. World Wide Web Site - Mouse Imprinting Data and References - http://www.har.mrc.ac.uk/research/genomic_imprinting
- ^
- ^
Nakka, P., Smith, S. P., O’Donnell-Luria, A. H., McManus, K. F., Agee, M., Auton, A., Bell, R. K., Bryc, K., Elson, S. L., Fontanillas, P., Furlotte, N. A., Hicks, B., Hinds, D. A., Jewett, E. M., Jiang, Y., Lin, K.-H., McCreight, J. C., Huber, K. E., Kleinman, A., … Sathirapongsasuti, J. F. (2019). Characterization of Prevalence and Health Consequences of Uniparental Disomy in Four Million Individuals from the General Population. The American Journal of Human Genetics, 105(5), 921–932. https://doi.org/10.1016/j.ajhg.2019.09.016
- ^
Robinson, W. P. (2000). Mechanisms leading to uniparental disomy and their clinical consequences. BioEssays, 22(5), 452–459. https://doi.org/10.1002/(SICI)1521-1878(200005)22:5<452::AID-BIES7>3.0.CO;2-K
- ^
UPD at conception is very unlikely to happen for most chromosomes except those chromosomes that most frequently segregate incorrectly, like the X-chromosome (which is aneuploid in ~ 0.5% of healthy sperm[15]). I couldn't find really good numbers on oocytes, but they tend to be aneuploid more frequently, somewhere between 3-10 times more often than sperm.
- ^
Spriggs, E. L., Rademaker, A. W., & Martin, R. H. (1996). Aneuploidy in human sperm: The use of multicolor FISH to test various theories of nondisjunction. American Journal of Human Genetics, 58(2), 356–362.
Discuss

