Published on January 27, 2025 10:00 AM GMT
This a very speculative thought and I have no expertise but, could it work?
Scanless Whole Brain Emulation attempts to simulate the development of the nervous system from the very start, thus not requiring a brain scan.
Hard, but necessary anyways?
For a Whole Brain Emulation to be useful, the simulated humans need to be able to learn. They need to understand new things and build on top of their new understandings, otherwise they would be intellectually disabled.
They need some level of neuroplasticity. The system needs to simulate how neurons use past information, as well how neurons form and prune connections. It needs to simulate the part of brain development in adulthood.
Given that it must simulate the part of brain development in adulthood, simulating brain development from the very start might not be that much harder.
But how would you even start?
The idea is to create protein models using chemistry simulations, and then create organelle models from protein simulations, and then create cell models from organelle simulations, and finally create "Scanless Whole Brain Emulation" from cell simulations.
Each model is created by running simulations of lower level mechanics, and fitting the simulation to empirical data. Once the simulation matches empirical data well enough, a model is then trained to match the simulation's behaviour.[1]
This differs from traditional Whole Brain Emulation, where the neuron models come from purely empirical data. Gaining empirical data about which neuron connects to which other neuron requires a brain scan, but brain scans are very hard to do.
By using lower level simulations, we can make a far finer model of the neuron, which captures all the important behaviours and computations that a neuron does.
A human cell such as a neuron has 750MB of data in its DNA or "source code", and may store a smaller amount of data in its epigenetics or "memory." It may store a much smaller amount of data in the concentrations of various molecules and ions, which is the very short term memory or "CPU buffer."
Its behaviour is very complex in the sense it is hard to understand. But it isn't very "computationally complex" in the sense the nonrandom and important computations are fewer than the computations in a cheap computer.
If we can model human neurons and cells accurately enough to capture most of their nonrandom and important computations, then we can not only simulate how they send signals, but simulate how they grow, move, and use past information (in their epigenetics).
Such a model would be useful for studying the mysterious mechanisms of many neurological diseases, and how interventions would influence the neurological disease, and this may be a better investment pitch than Whole Brain Emulation.
But if the neurons and cells can be modelled both accurately and cheaply, it may be possible to simulate the development of the nervous system from the very start.
Details
- The FHI's article on Whole Brain Emulation lists the Simulation Scales which a simulation could occur on. They are not talking about this idea of Scanless Whole Brain EmulationIn order to fit simulations to empirical data, or at least verify that simulations fit empirical data, it may be best to start with small animals like roundworms. Roundworms have already been simulated. It's important to start with smaller animals and gradually move up to humans, to make sure the simulated beings are not suffering in unnatural ways. Fit the embryo simulations to empirical data before simulating later development. An unhappy simulated human may be as bad as a misaligned AI.I do not expect a human effort on this project to succeed faster than a human effort to build superintelligence (without Whole Brain Emulation). The only way it'll work is if superintelligence efforts are paused for a long time.
- Or if superintelligence is paused for a short time, and human-level AGI (which is good at engineering and thinks many times faster) can build it within that short time. But this sounds like a far shot.If the system is built, converting the simulated human into a human-like superintelligence should be relatively easier than building it in the first place.
AI can simulate physics and other systems far more cheaply than traditional physics simulators
A random example is the AI simulating the exact behaviour of potassium atoms, and correctly predicting the existence of a new state of matter.
"Why didn't they use traditional physics simulators without AI?" Well, to match the behaviour of real atoms accurately enough, you must take quantum mechanics into account. Simulating atoms in a way which matches quantum mechanics, gets exponentially harder with more atoms (on a non-quantum computer).
AI on the other hand, learns a bunch of heuristics which approximates how quantum mechanics behaves, without doing the really hard quantum computations. How exactly it succeeds is mysterious, it just does.
Here is the beautiful video that I learned this from:
The mechanics of a bunch of potassium atoms is relatively simple, but even more complex phenomena like protein folding is being predicted better and better by Google DeepMind's AlphaFold.
What's amazing about AlphaFold is that people tried many machine learning approaches on the protein folding problem for a long time. There was this competition called the Critical Assessment of Structure Prediction, where most attempts didn't getting very close to accurate results. AlphaFold scored thrillingly better than the other methods in a way no one thought possible.
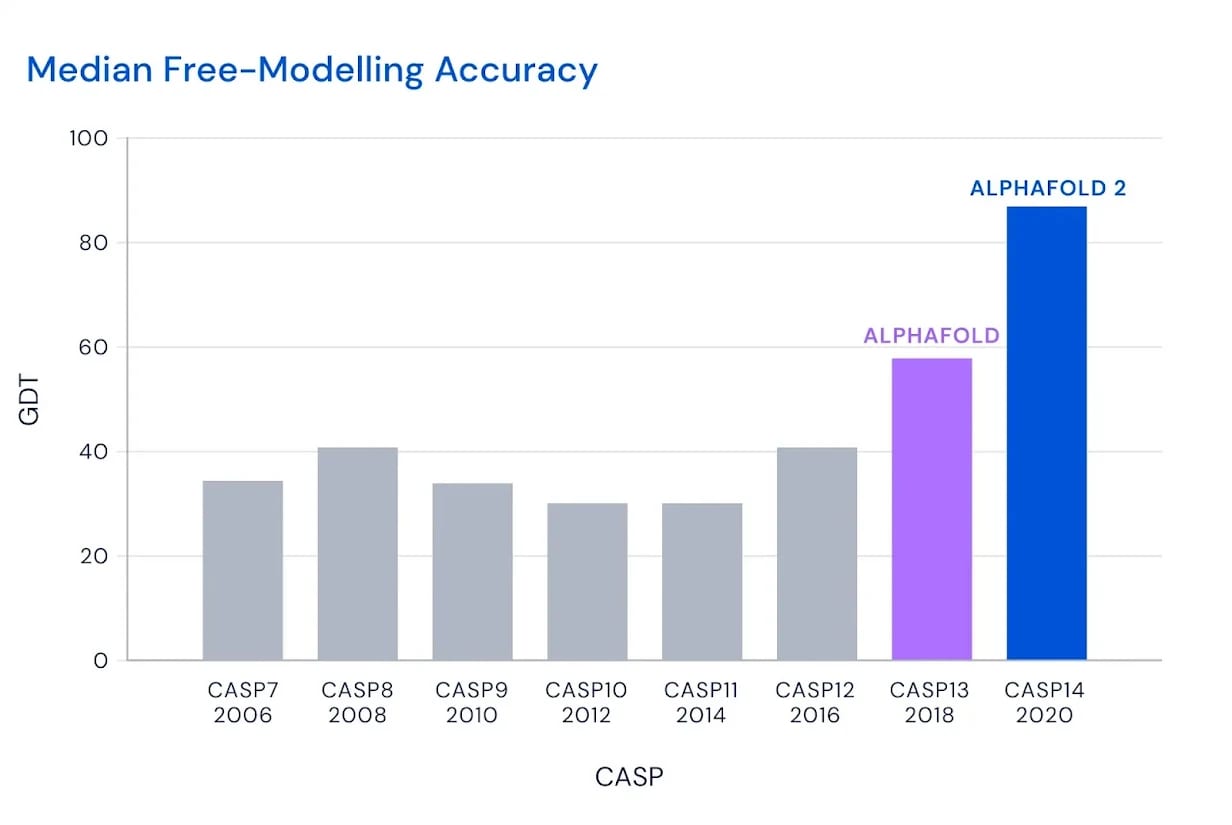
It's an open question what other breakthroughs can be made in the field of predicting biochemistry with AI.
- ^
There are analogies to Iterated Amplification.
Discuss

