Drug discovery is a complex, time-intensive process that requires researchers to navigate vast amounts of scientific literature, clinical trial data, and molecular databases. Life science customers like Genentech and AstraZeneca are using AI agents and other generative AI tools to increase the speed of scientific discovery. Builders at these organizations are already using the fully managed features of Amazon Bedrock to quickly deploy domain-specific workflows for a variety of use cases, from early drug target identification to healthcare provider engagement.
However, more complex use cases might benefit from using the open source Strands Agents SDK. Strands Agents takes a model-driven approach to develop and run AI agents. It works with most model providers, including custom and internal large language model (LLM) gateways, and agents can be deployed where you would host a Python application.
In this post, we demonstrate how to create a powerful research assistant for drug discovery using Strands Agents and Amazon Bedrock. This AI assistant can search multiple scientific databases simultaneously using the Model Context Protocol (MCP), synthesize its findings, and generate comprehensive reports on drug targets, disease mechanisms, and therapeutic areas. This assistant is available as an example in the open-source healthcare and life sciences agent toolkit for you to use and adapt.
Solution overview
This solution uses Strands Agents to connect high-performing foundation models (FMs) with common life science data sources like arXiv, PubMed, and ChEMBL. It demonstrates how to quickly create MCP servers to query data and view the results in a conversational interface.
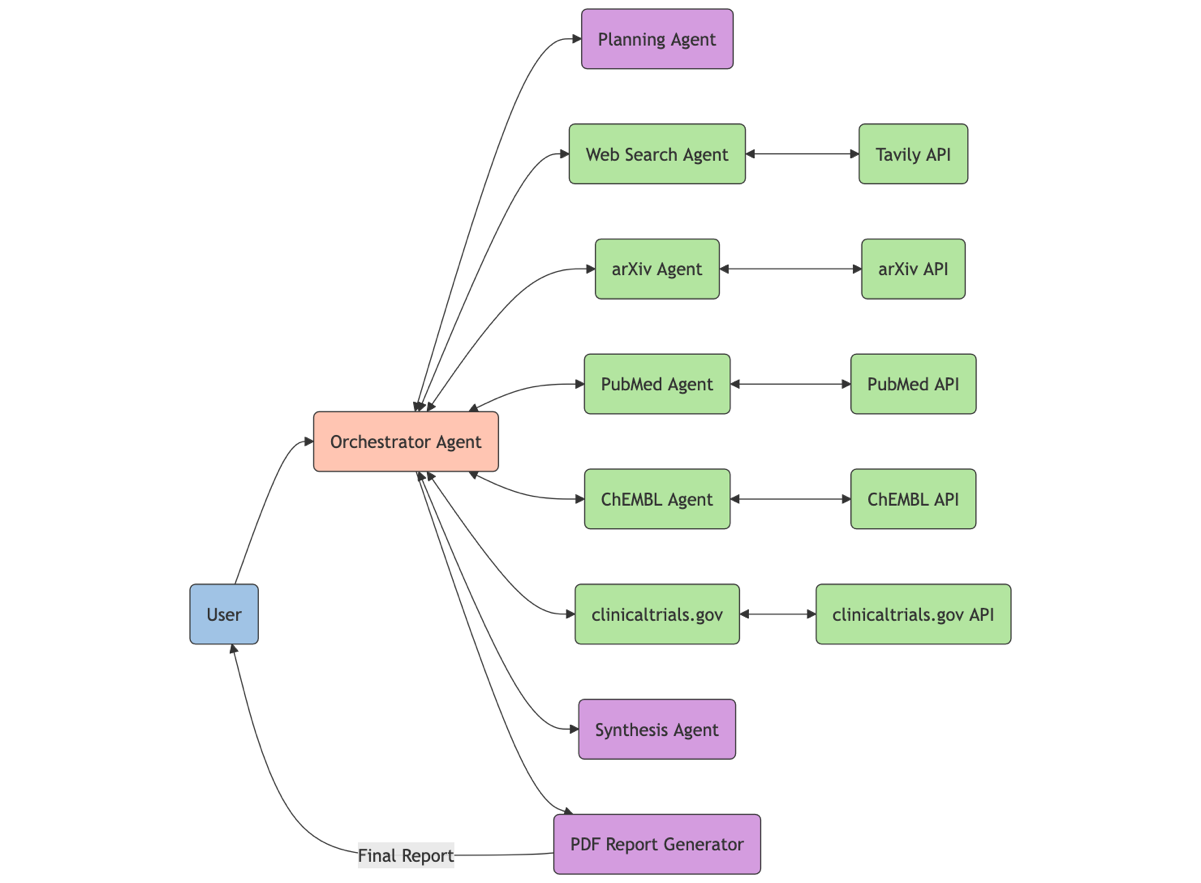
Small, focused AI agents that work together can often produce better results than a single, monolithic agent. This solution uses a team of sub-agents, each with their own FM, instructions, and tools. The following flowchart shows how the orchestrator agent (shown in orange) handles user queries and routes them to sub-agents for either information retrieval (green) or planning, synthesis, and report generation (purple).
This post focuses on building with Strands Agents in your local development environment. Refer to the Strands Agents documentation to deploy production agents on AWS Lambda, AWS Fargate, Amazon Elastic Kubernetes Service (Amazon EKS), or Amazon Elastic Compute Cloud (Amazon EC2).
In the following sections, we show how to create the research assistant in Strands Agents by defining an FM, MCP tools, and sub-agents.
Prerequisites
This solution requires Python 3.10+, strands-agents, and several additional Python packages. We strongly recommend using a virtual environment like venv or uv to manage these dependencies.
Complete the following steps to deploy the solution to your local environment:
- Clone the code repository from GitHub. Install the required Python dependencies with
pip install -r requirements.txt. Configure your AWS credentials by setting them as environment variables, adding them to a credentials file, or following another supported process. Save your Tavily API key to a .env file in the following format: TAVILY_API_KEY="YOUR_API_KEY". You also need access to the following Amazon Bedrock FMs in your AWS account:
- Anthropic’s Claude 3.7 Sonnet Anthropic’s Claude 3.5 Sonnet Anthropic’s Claude 3.5 Haiku
Define the foundation model
We start by defining a connection to an FM in Amazon Bedrock using the Strands Agents BedrockModel class. We use Anthropic’s Claude 3.7 Sonnet as the default model. See the following code:
Define MCP tools
MCP provides a standard for how AI applications interact with their external environments. Thousands of MCP servers already exist, including those for life science tools and datasets. This solution provides example MCP servers for:
- arXiv – Open-access repository of scholarly articles PubMed – Peer-reviewed citations for biomedical literature ChEMBL – Curated database of bioactive molecules with drug-like properties ClinicalTrials.gov – US government database of clinical research studies Tavily Web Search – API to find recent news and other content from the public internet
Strands Agents streamlines the definition of MCP clients for our agent. In this example, you connect to each tool using standard I/O. However, Strands Agents also supports remote MCP servers with Streamable-HTTP Events transport. See the following code:
Define specialized sub-agents
The planning agent looks at user questions and creates a plan for which sub-agents and tools to use:
Similarly, the synthesis agent integrates findings from multiple sources into a single, comprehensive report:
Define the orchestration agent
We also define an orchestration agent to coordinate the entire research workflow. This agent uses the SlidingWindowConversationManager class from Strands Agents to store the last 10 messages in the conversation. See the following code:
Example use case: Explore recent breast cancer research
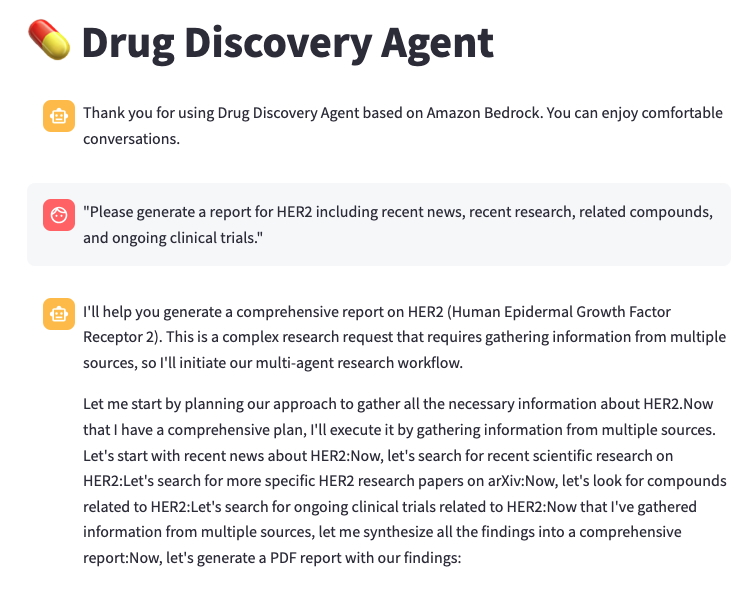
To test out the new assistant, launch the chat interface by running streamlit run application/app.py and opening the local URL (typically http://localhost:8501) in your web browser. The following screenshot shows a typical conversation with the research agent. In this example, we ask the assistant, “Please generate a report for HER2 including recent news, recent research, related compounds, and ongoing clinical trials.” The assistant first develops a comprehensive research plan using the various tools at its disposal. It decides to start with a web search for recent news about HER2, as well as scientific articles on PubMed and arXiv. It also looks at HER2-related compounds in ChEMBL and ongoing clinical trials. It synthesizes these results into a single report and generates an output file of its findings, including citations.
The following is an excerpt of a generated report:
Notably, you don’t have to define a step-by-step process to accomplish this task. By providing the assistant with a well-documented list of tools, it can decide which to use and in what order.
Clean up
If you followed this example on your local computer, you will not create new resources in your AWS account that you need to clean up. If you deployed the research assistant using one of those services, refer to the relevant service documentation for cleanup instructions.
Conclusion
In this post, we showed how Strands Agents streamlines the creation of powerful, domain-specific AI assistants. We encourage you to try this solution with your own research questions and extend it with new scientific tools. The combination of Strands Agents’s orchestration capabilities, streaming responses, and flexible configuration with the powerful language models of Amazon Bedrock creates a new paradigm for AI-assisted research. As the volume of scientific information continues to grow exponentially, frameworks like Strands Agents will become essential tools for drug discovery.
To learn more about building intelligent agents with Strands Agents, refer to Introducing Strands Agents, an Open Source AI Agents SDK, Strands Agents SDK, and the GitHub repository. You can also find more sample agents for healthcare and life sciences built on Amazon Bedrock.
For more information about implementing AI-powered solutions for drug discovery on AWS, visit us at AWS for Life Sciences.
About the authors
 Hasun Yu is an AI/ML Specialist Solutions Architect with extensive expertise in designing, developing, and deploying AI/ML solutions for healthcare and life sciences. He supports the adoption of advanced AWS AI/ML services, including generative and agentic AI.
Hasun Yu is an AI/ML Specialist Solutions Architect with extensive expertise in designing, developing, and deploying AI/ML solutions for healthcare and life sciences. He supports the adoption of advanced AWS AI/ML services, including generative and agentic AI.
 Brian Loyal is a Principal AI/ML Solutions Architect in the Global Healthcare and Life Sciences team at Amazon Web Services. He has more than 20 years’ experience in biotechnology and machine learning and is passionate about using AI to improve human health and well-being.
Brian Loyal is a Principal AI/ML Solutions Architect in the Global Healthcare and Life Sciences team at Amazon Web Services. He has more than 20 years’ experience in biotechnology and machine learning and is passionate about using AI to improve human health and well-being.